It’s been a busy semester! I’m studying in Durham, North Carolina under the NESCent graduate fellowship, where daily I grow fonder of the city’s rugged tobacco-and-cinder aesthetic. Not so fond that I’ve forgotten Oakland, of course.
While here, part of my plan is to explore new biogeographic range evolution models designed for large numbers of areas (see initial work here). The data augmentation method I’m using was originally introduced to phylogenetics for use with protein evolution by Robinson et al. (2003) from Jeff Thorne’s group and others. To my great fortune, being a short bus ride away to Raleigh from Durham, Jeff generously agreed to act as my research advisor for the semester.
In early September, I presented some past work on applying Lévy processes to phylogenetic inference (collaboration with Josh Schraiber and Mason Liang) at the Mathematics for an Evolving Biodiversity workshop hosted at the University of Montréal. A very stimulating conference! Mid-September, I presented on Bayesian biogeography for the Phylogenetics & Evolutionary Biology Seminar group at NCSU.
With the presentations aside, I’ve had time to focus more on my fellowship project. With a little effort, I now have a working version of BayArea implemented in RevBayes. It’s extremely easy to tinker with new dispersal models in this framework, so I expect my next biogeography inference method will be released solely under RevBayes.
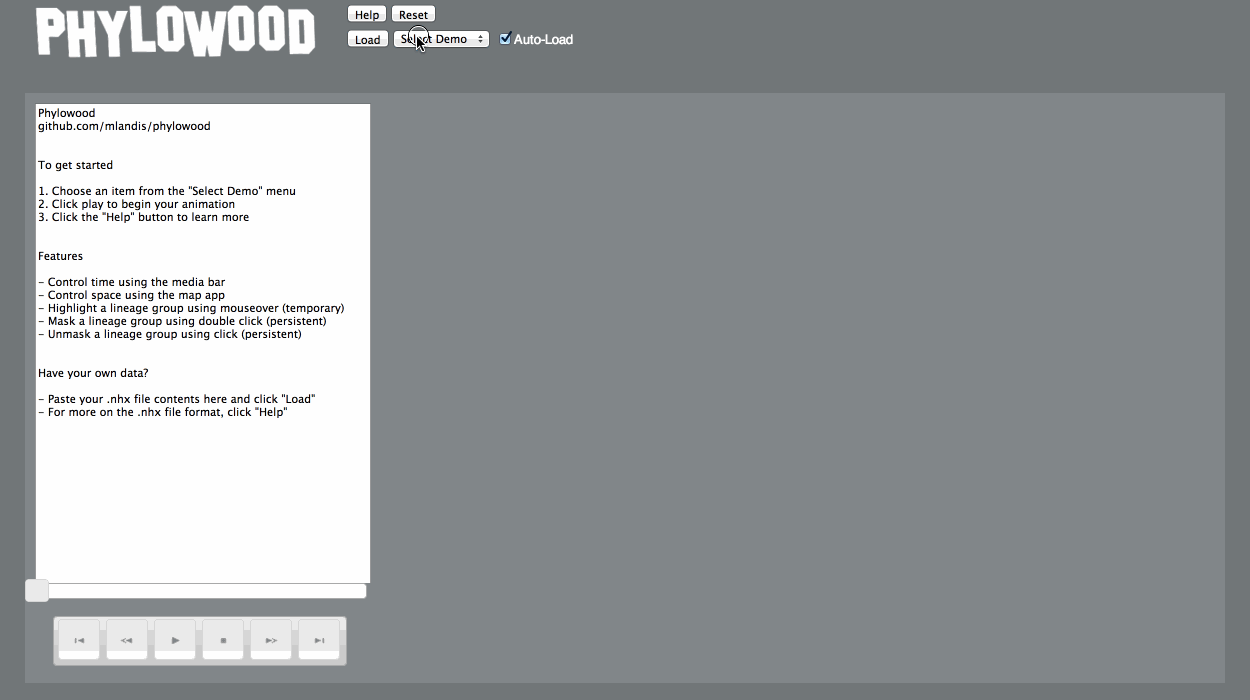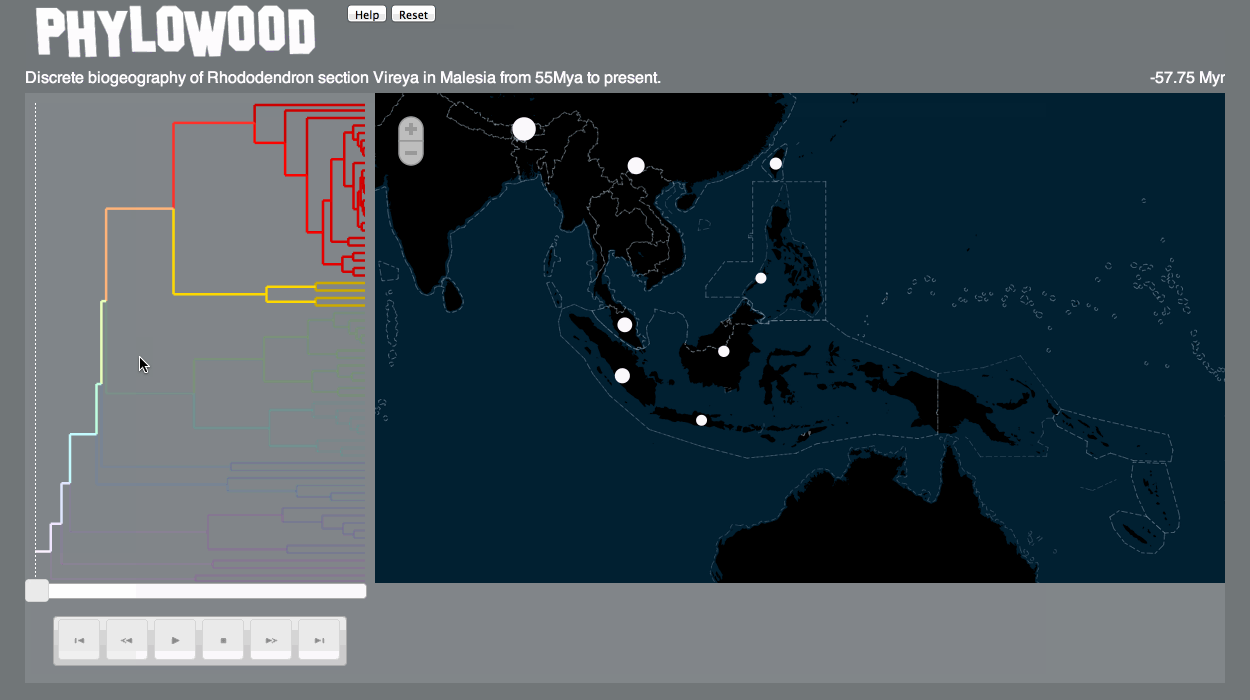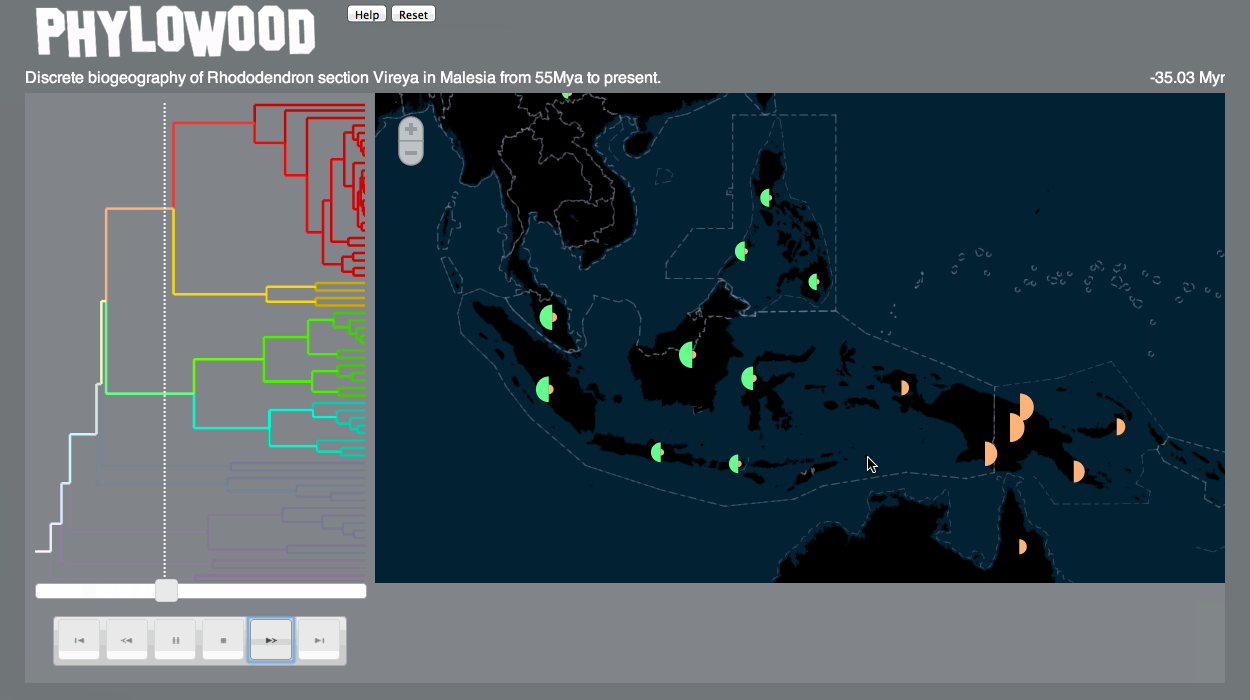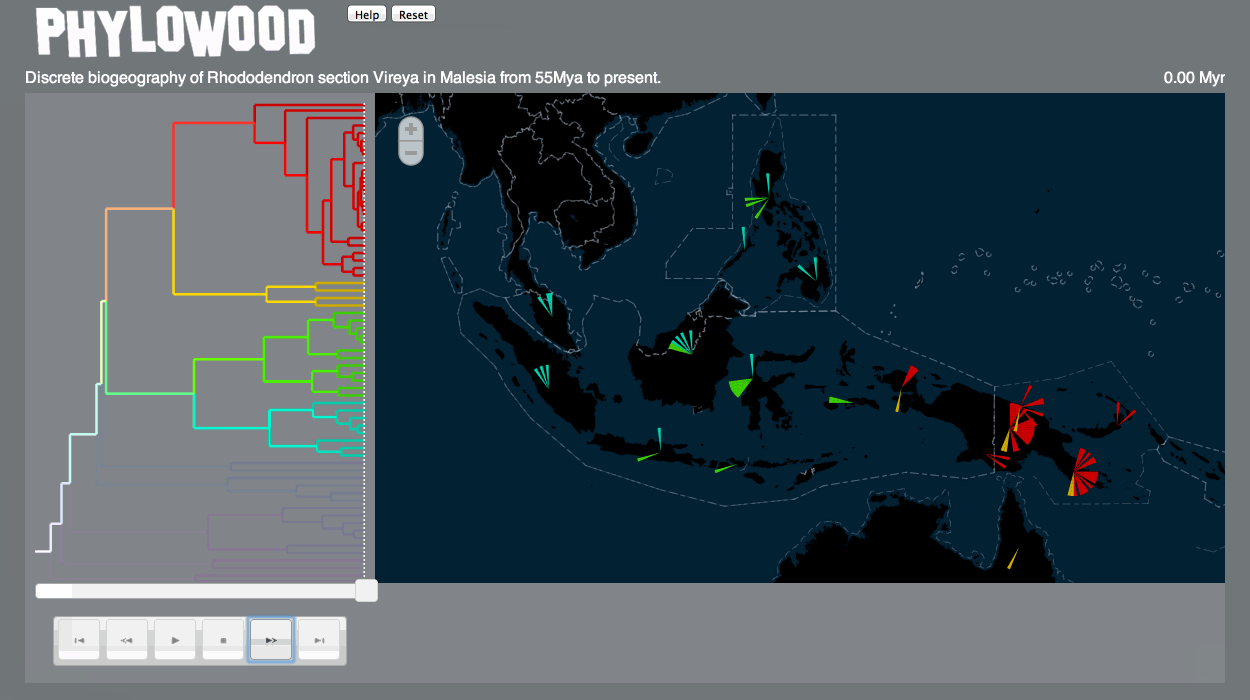
Finally, I’m happy to say that Phylowood – what I began on a bit of a whim as a “fun and relaxing” post-qualifying exam project, then matured into a Google Summer of Code project – is now published as a Bioinformatics Application Note (link). Trevor Bedford, my GSoC mentor, was very supportive throughout the whole process, and even covered the Open Access fee.
Here’s a gif demonstrating some of the features of Phylowood:
Plenty in the pipeline to come, but that’s all to report on for now!